Output page
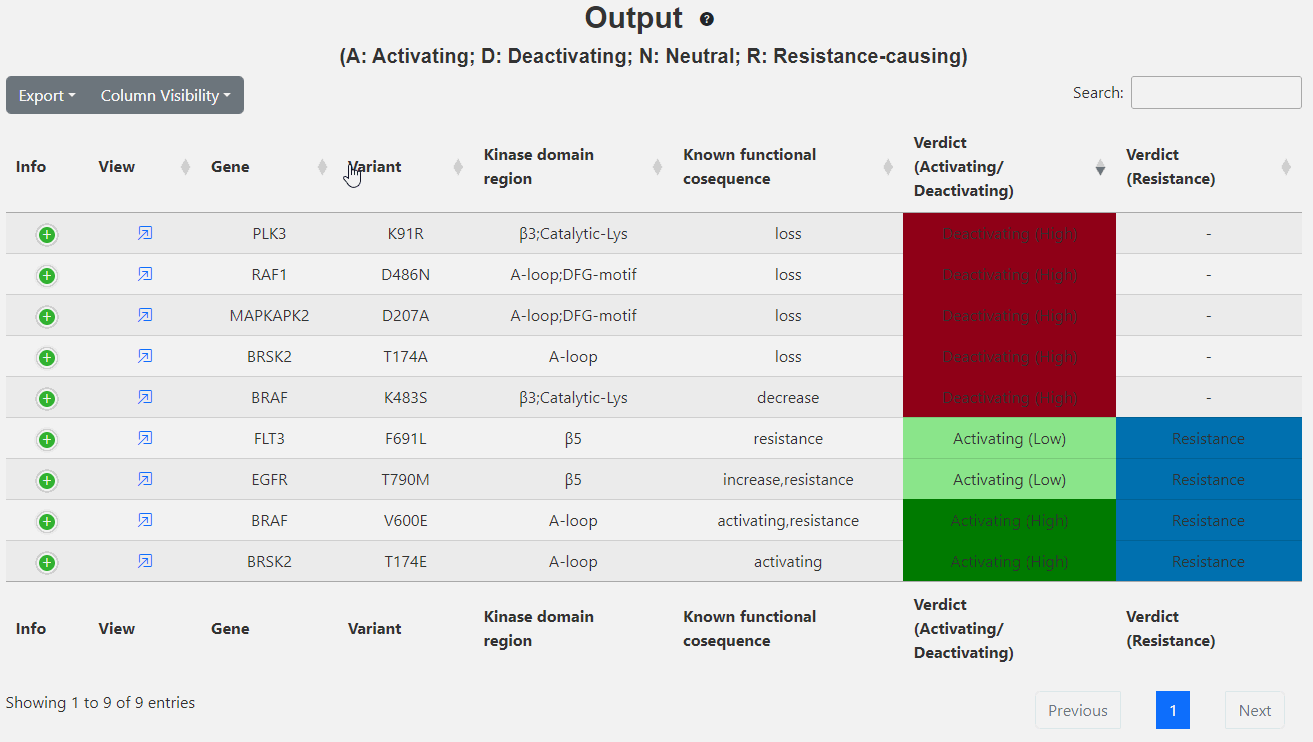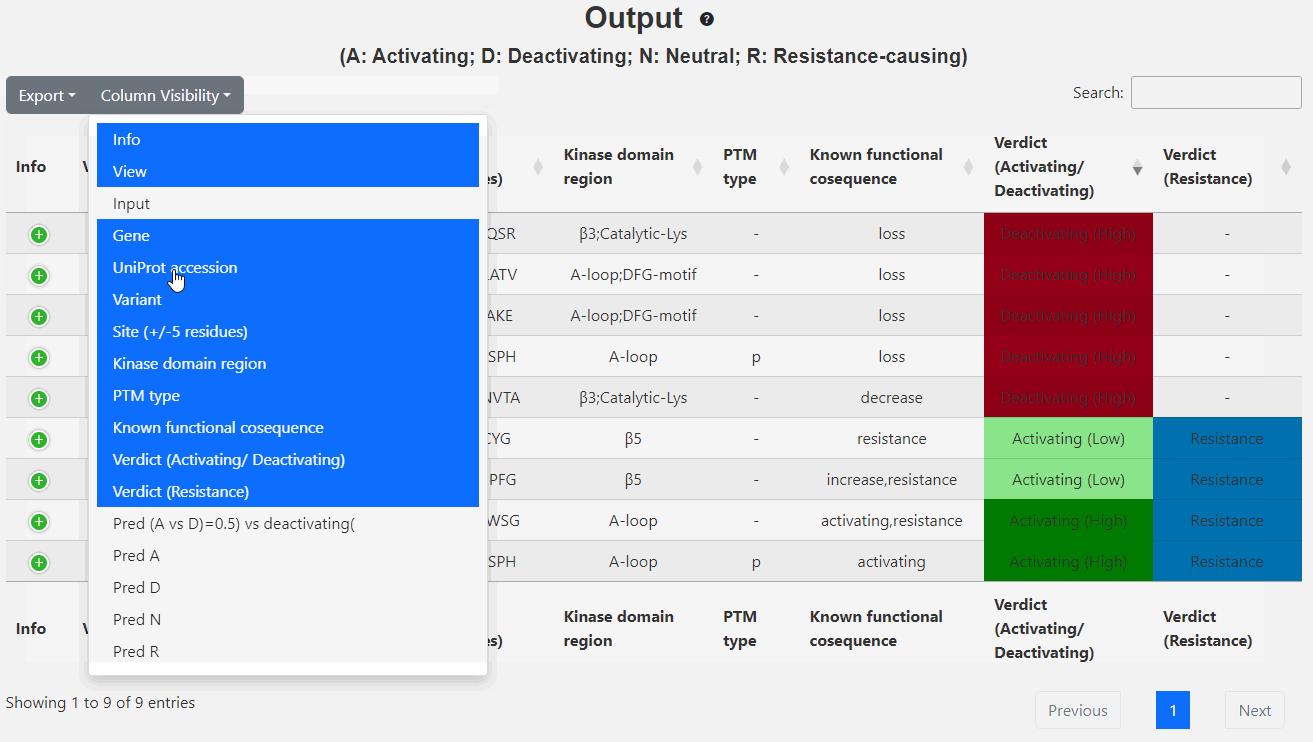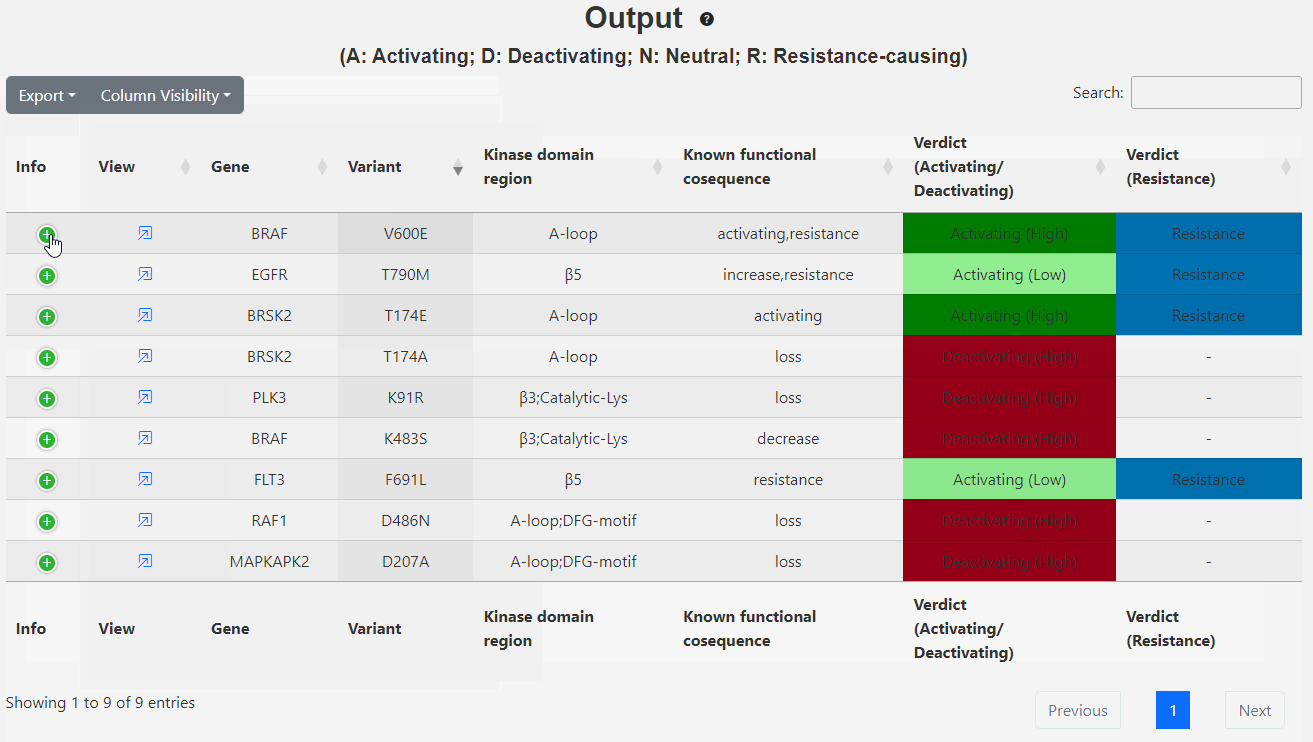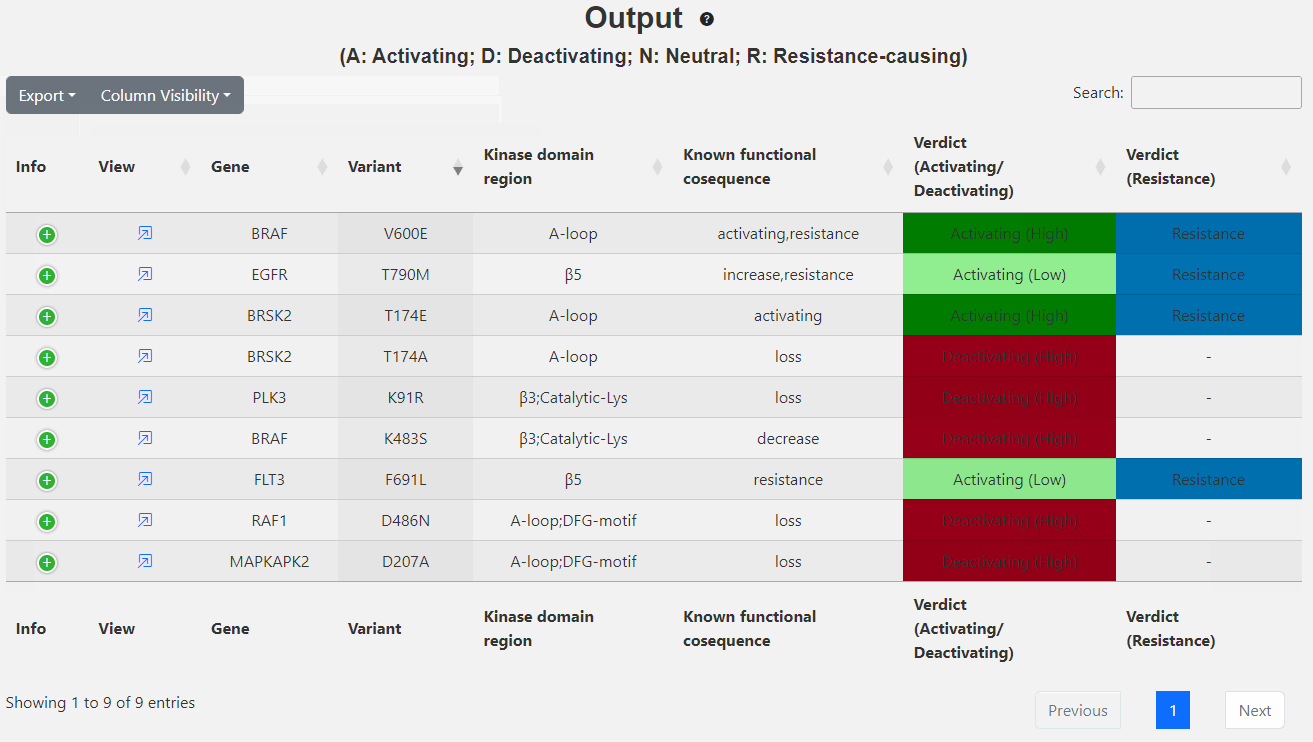
This page displays the brief output of the inputs:
- Info: Known functional and PTM information at the equivalent site in other kinases.
- View: Click to view more descrition of the variant in another tab.
- Gene: Gene name of the kinase.
- Variant: Input variant.
- Kinase domain region: Kinase domain region.
- Known functional consequence: Any known functional consequence of the input variant.
- Verdict (activating vs deactivating): Likelihood if the given variant is activating or deactivating the kinase. Values in paranthesis refer to the prediction confidence (Low, Medium, High). Variants with uncertain verdicts are marked as "ambigous" and the ones that lie outside the kinase domain as '-'.
- Verdict (resistance): Likelihood if the given variant is resistant the kinase. Values in paranthesis refer to the prediction confidence (Low, Medium. High). Variants that lie outside the kinase domain as '-'.
Summary of the variant
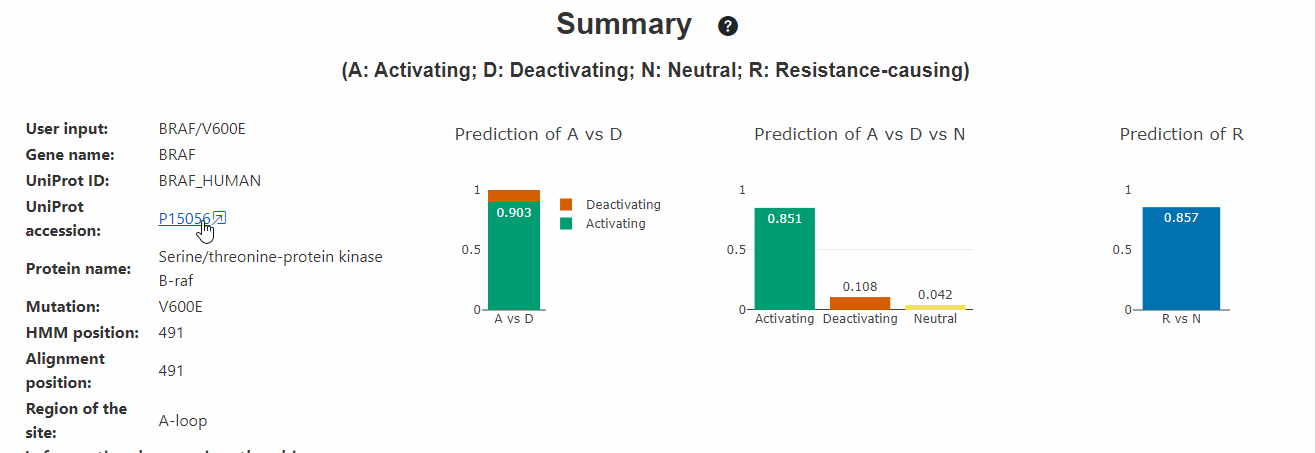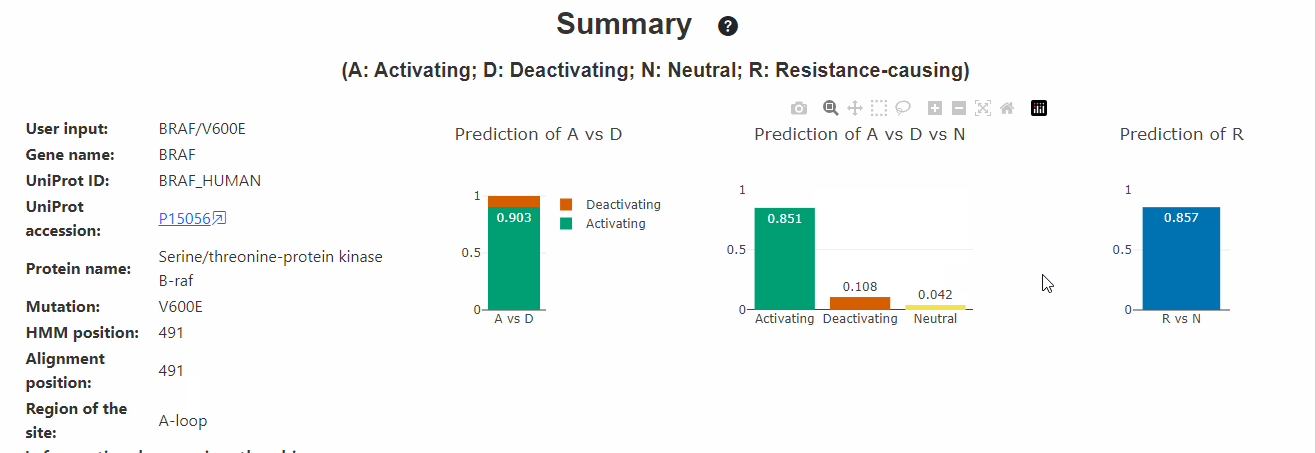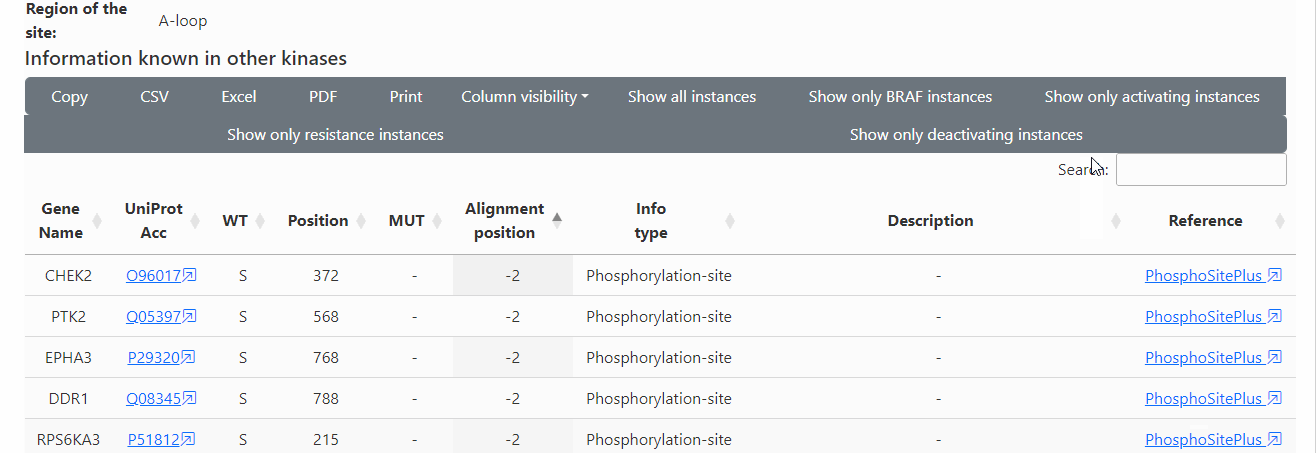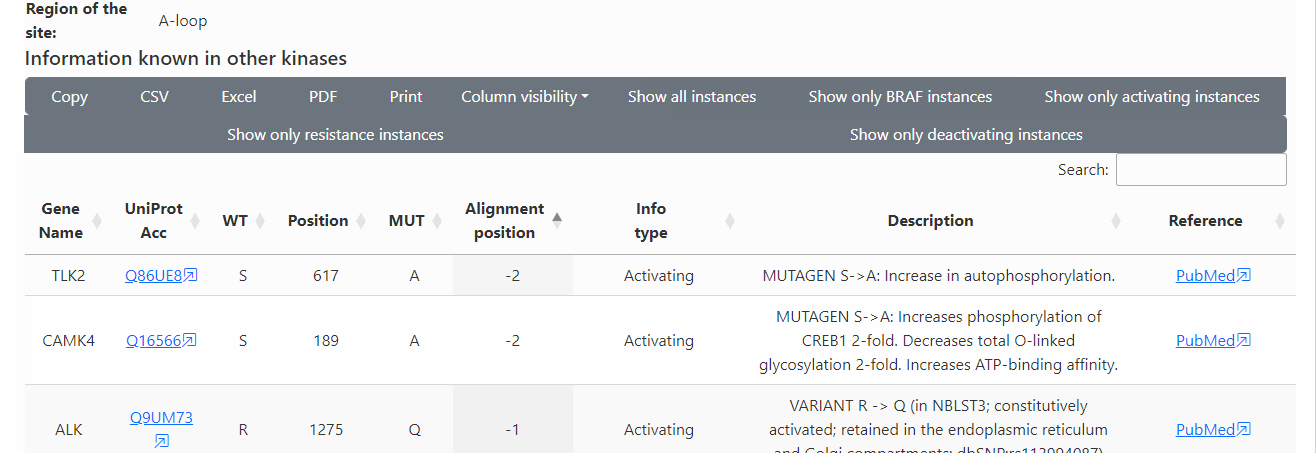
This section displays the summary of the input variant, predicted probabilities and known functional and PTM information of the variant site in all kinases. On the left side of the section, brief summary of the input is displayed. On the right side, the following 3 plots are displayed:
- Prediction (Activating vs Deactivating): These predictions represent a typical situation when one has what is believed to be a functional variant (e.g. observed many times in a cohort or dataset) and wishes to distinguish these two possibilities.
- Prediction (Activating vs Deactivating vs Neutral): These predictions are more reflective of a situation where one does not know if a variant is functional at all and thus one needs to predict neutrals.
- Prediction (Resistance vs Not): This predicts if a given variant is resistant or not. Note: Given that this predictor lacks information about activating and deactivating information, users may ignore its predictions when other predictors predict the variant to be strongly deactivating (eg: variants occuring at most conserved residues like the Catalytic Lysine, DFG-motif, etc., which are associated with loss of kinase activity.)
Known functional and PTM information in all kinases: The table below (scroll) shows the known functional and PTM information of the variant site and adjacent positions (+/- 2 in the alignment) in all kinases.
Alignment
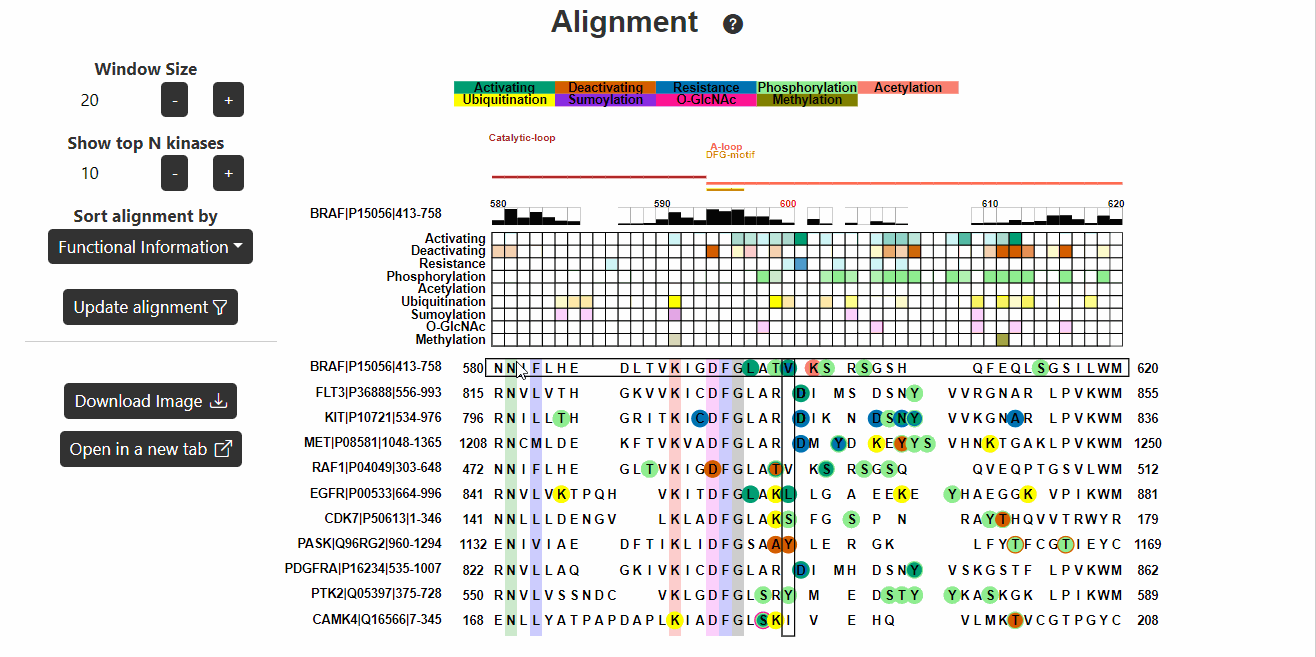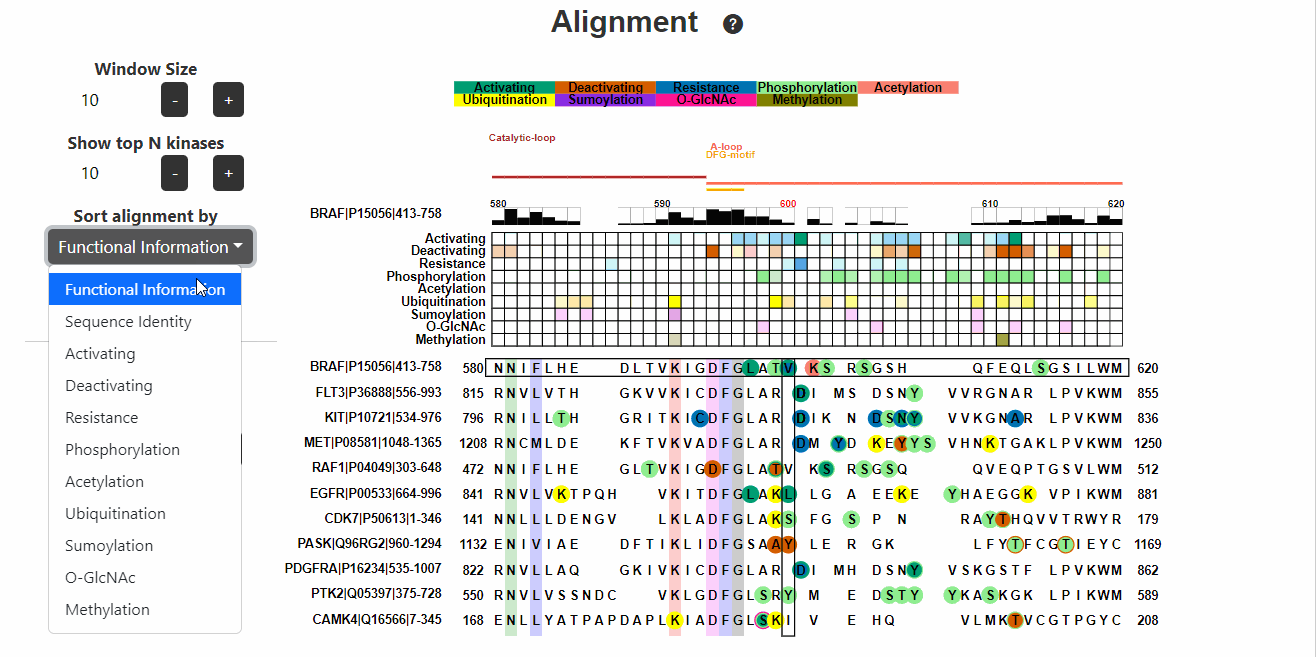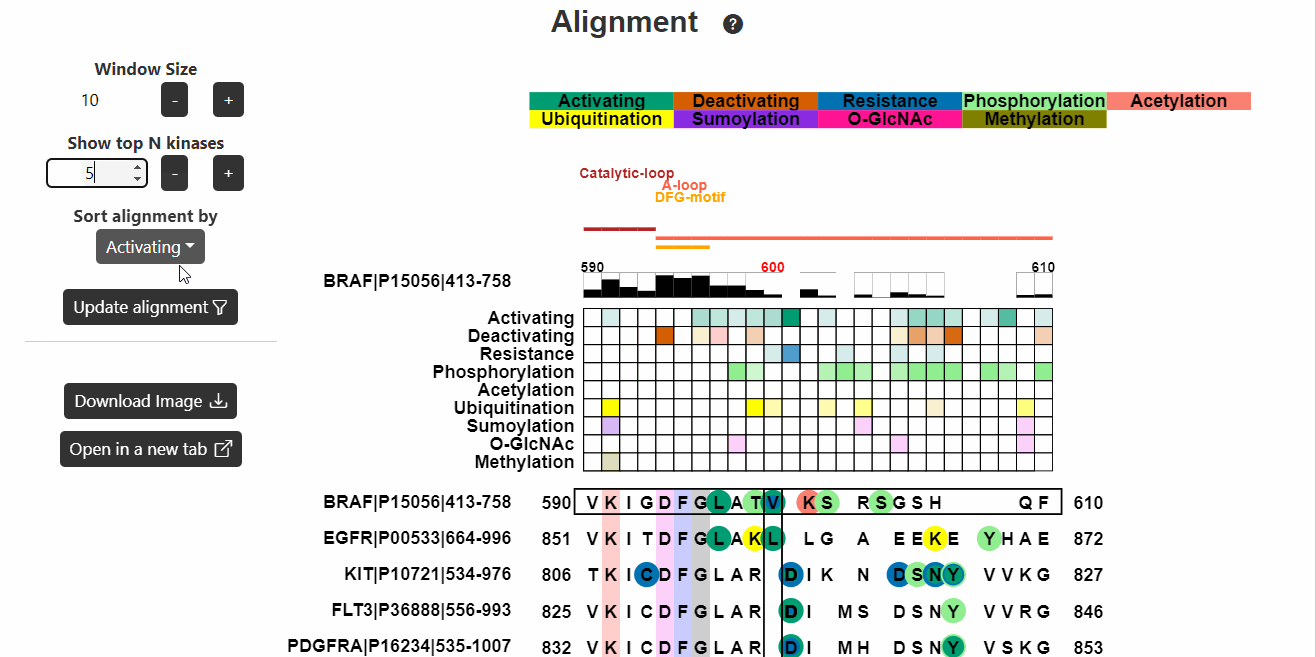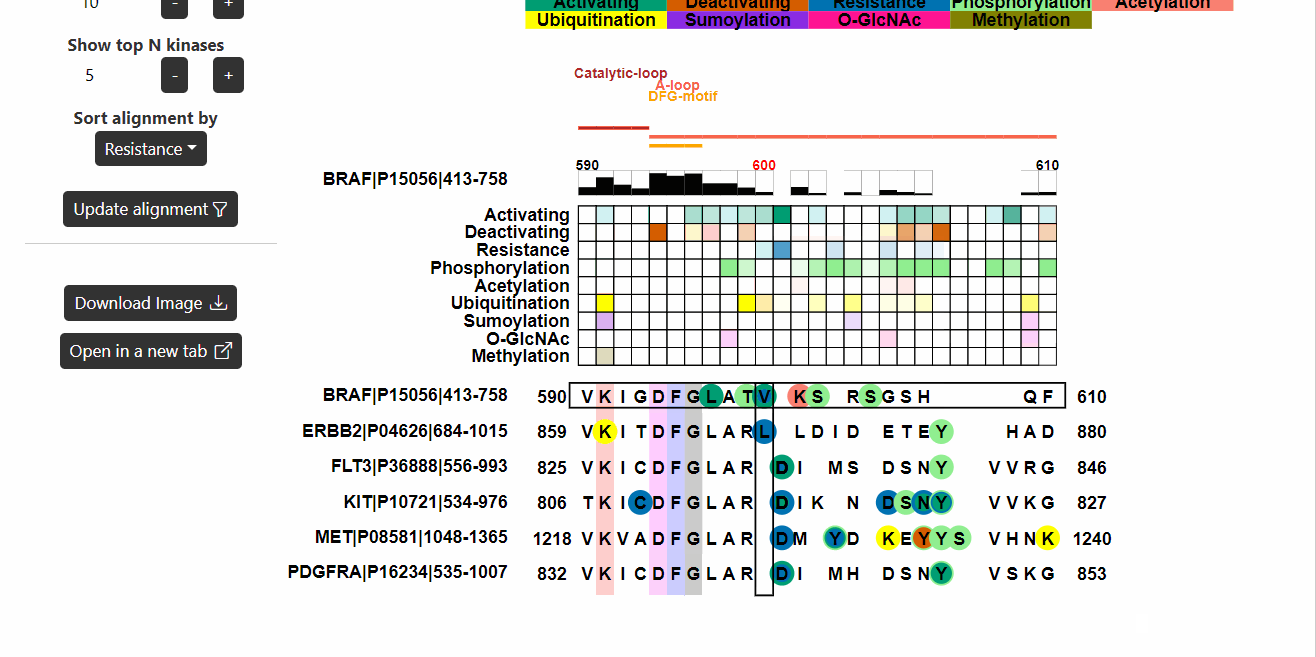
This section displays the alignment of the input kinase sequence with
other human kinase sequences. Known functional variants are then mapped
onto the alignment. Hover on the variants to know more about them.
- Window Size represents the number of positions before or after the variant site to be displayed in the alignment (recommended 10-30).
- Show top N kinases reflects the top N kinases to be displayed in the alignment. (recommended 10-100).
- Sort alignment by represents the information (known functional or PTM) based on which the alignment will be sorted.
Contacts
Gurdeep Singh gurdeep.singh[at]bioquant[dot]uni-heidelberg[dot]deTorsten Schmenger torsten.schmenger[at]bioquant[dot]uni-heidelberg[dot]de
Rob Russell robert.russell[at]bioquant[dot]uni-heidelberg[dot]de