When to use the resistant or not predictor?
This predictor predicts if the given variant is resistant or not.
However, it does not take into account if the variant is activating or deactivating. Hence,
this predictor is useful when one knows that a variant DOES NOT lead to loss of
kinase activity but one does not know if the variant is resistant or not (see example below).
For example:
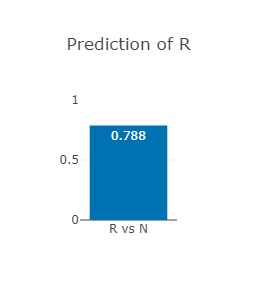
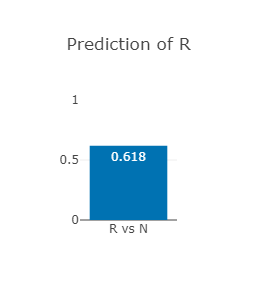
The EGFR p.Thr790Met variant
Thr790 lies in the β5 sheet of EGFR
Thr790Met is predicted to be drug-resistant (see right).
Note that this predictor predcits the likelihood of
a given variant to be resistant to a drug, not the likelihood
of a patient to respond to a drug.
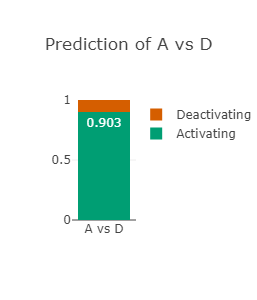
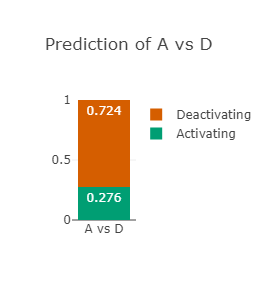
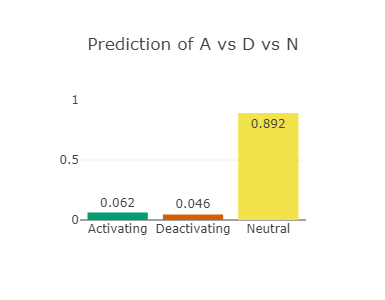
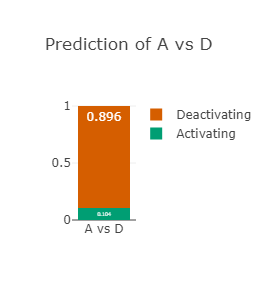
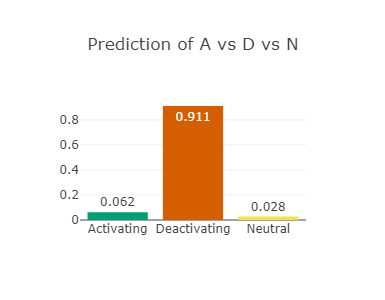
The RAF1 p.Asp486Asn variant (CAUTION)
Asp486 lies in the activation-loop and at the DFG-motif in RAF1
Asp486 is predicted to be drug-resistant (see right-most figure).
Asp486 is also predicted to be strongly deactivating (see left-most and middle figures)
Since the resistance or not predictor lacks the information about activating or deactivating,
it is not able to take this into account and
hence the prediction of drug-resistance should be ignored.